Abstract
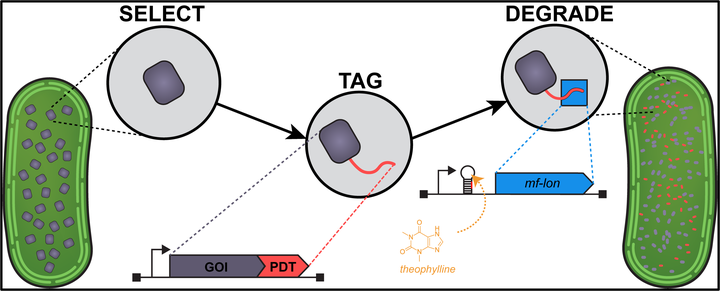
Synechococcus elongatus PCC 7942 is a model cyanobacterium for study of the circadian clock, photosynthesis, and bioproduction of chemicals, yet nearly 40% of its gene identities and functions remain unknown, in part due to limitations of the existing genetic toolkit. While classical techniques for the study of genes (e.g., deletion or mutagenesis) can yield valuable information about the absence of a gene and its associated protein,there are limits to these approaches, particularly in the study of essential genes. Herein, we developed a tool for inducibledegradation of target proteins in S. elongatus by adapting a method using degron tags from the Mesoplasma florum transfer-mRNA (tmRNA) system. We observed that M. florum lon protease can rapidly degrade exogenous and native proteins tagged with the cognate sequence within hours of induction. We used this system to inducibly degrade the essential cell division factor, FtsZ, as well as shell protein components of the carboxysome. Our results have implications for carboxysome biogenesis and the rate of carboxysome turnover during cell growth. Lon protease control of proteins offers an alternative approach for the study of essential proteins and protein dynamics in cyanobacteria.